Did you know that over 300 million people worldwide live with a rare disease? A "rare disease" is classified as occurring in fewer than 1 in 2,000 people, but collectively, with over 7,000 distinct rare genetic diseases identified, they are common. Rare disease patients and their families can face painful diagnostic odysseys, but trio sequencing can help.
The Unique Challenge of Rare Disease
Families impacted by rare diseases can face tremendous challenges getting help. Symptoms typically manifest in early childhood, are frequently severe, and are often nonspecific or obscure. By definition, their rarity means a typical doctor will not encounter more than one case of a given disease in their entire career. A patient's diagnostic odyssey can last years and include multiple specialists, tests, invasive procedures, and misdiagnoses. The emotional and financial toll on the patient and their family is very high.
In recent years, whole exome sequencing (WES) and whole genome sequencing (WGS) have offered families a shortcut to this diagnostic odyssey by identifying the critical genetic mutation that causes disease.2,3 The success rate of WGS can be improved by sequencing the parents alongside the patient.4 Trio sequencing identifies inheritance patterns, revealing cases where both parents are heterozygous carriers, and the child is homozygous. It is also more effective at uncovering de novo variants arising exclusively in the child, as the parental genomes can be used as healthy controls. Since every individual carries between 40,000-200,000 rare variants present in less than 0.5% of the population but only ~70 de novo variants, having parental genomes can significantly reduce the list of candidate causal genes that must be reviewed. The downside of trio sequencing is that with triple the genomes to sequence, there's triple the cost.
A New Approach to Trio Sequencing
The Element AVITI™ System, our mid-throughput DNA sequencing platform, offers a new way to do trio sequencing while lowering costs. In proof-of-concept work, we aimed to demonstrate that a complete trio study on the AVITI benchtop system can be performed for only $1,680 in sequencing reagents.
Typically, WGS sequencing requires 35x coverage (360 M read pairs) to generate robust variant calling data. That translates into approximately 1 billion read pairs for a trio – exactly matching the recently upgraded AVITI performance specification. However, our partners at Google DeepVariant recently published a manuscript showing that parental coverage can be reduced in trio sequencing while maintaining high variant calling accuracy and excellent de novo variant calling capability.5
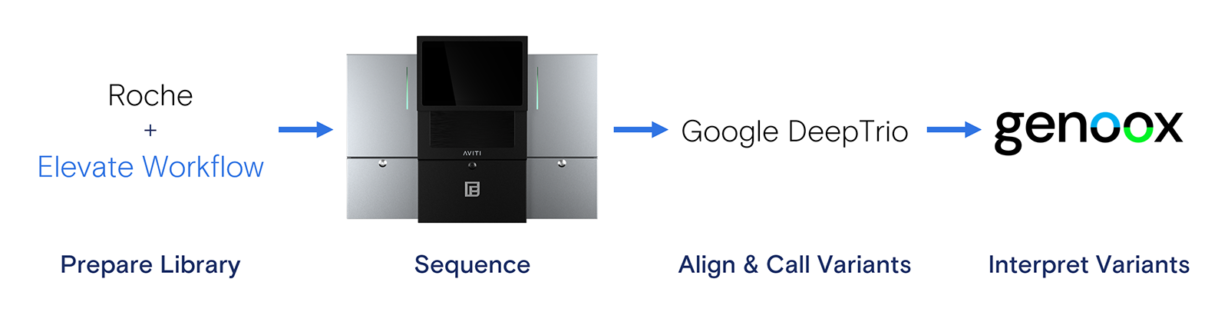
To add robustness to the experimental design, we decided to test pooling the trio samples such that the parent library concentrations were half that of the child library. We outlined a complete trio sequencing workflow working with leaders in next-generation sequencing (NGS) for rare disease research. We paired the Roche HyperPlus kit with the Element Elevate™ Library Preparation Workflow for library prep and selected Google DeepTrio for variant calling, following reference alignment via BWA-MEM. The DeepVariant team has developed an Element-optimized version of their neural network-based algorithm. Genoox Franklin was selected for variant interpretation and reporting. Genoox provides an easy-to-use graphical user interface that includes variant annotation and supporting publications, allowing you to easily connect with other researchers in your community with the same disease focus. DeepTrio and Genoox are widely used in leading institutions, providing low or no-cost options for their analysis pipelines. The complete sequencing and analysis workflow is shown in Figure 1.
Partnering with UC San Diego to Find the Drivers of Rare Eye Disease
We partnered with rare eye disease experts UC San Diego Professor Radha Ayyagari, PhD, and her graduate student, Pooja Biswas, to demonstrate the potential of this application to reduce costs without sacrificing efficacy.
You may recall that we previously worked with Professor Ayyagari's work was presented at our launch event, where Element sequenced DNA on the AVITI system from 10 subjects known to suffer from retinitis pigmentosa. We leveraged partnerships with Roche, Sentieon, Genoox, and Fabric to identify high-confidence candidates in five cases—three of which were previously unsolved. Pending clinical validation, this represents a 50% solve rate. Watch the video to learn more about the specific variants that were discovered.
For this new collaboration, we took up yet another rare eye disease case where none of the family had previously been sequenced, aiming to explore what WGS of the full mother, father, and child trio might reveal, using our newly outlined workflow.
New Hope for an Answer
The run generated over 1.1 billion read pairs and 326 GB of sequencing data, yielding greater than 20x in each parent and 50x in the proband, with 94.6% of the data at Q30 of higher.
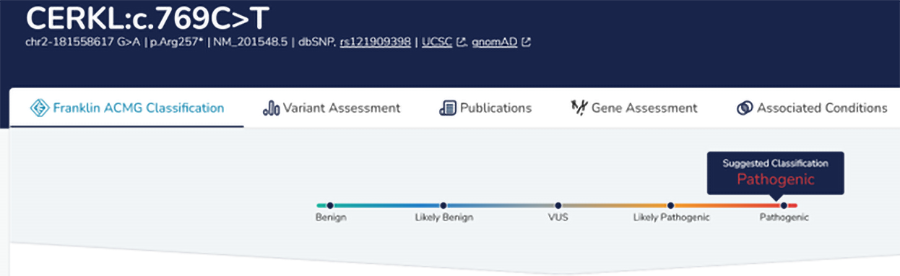
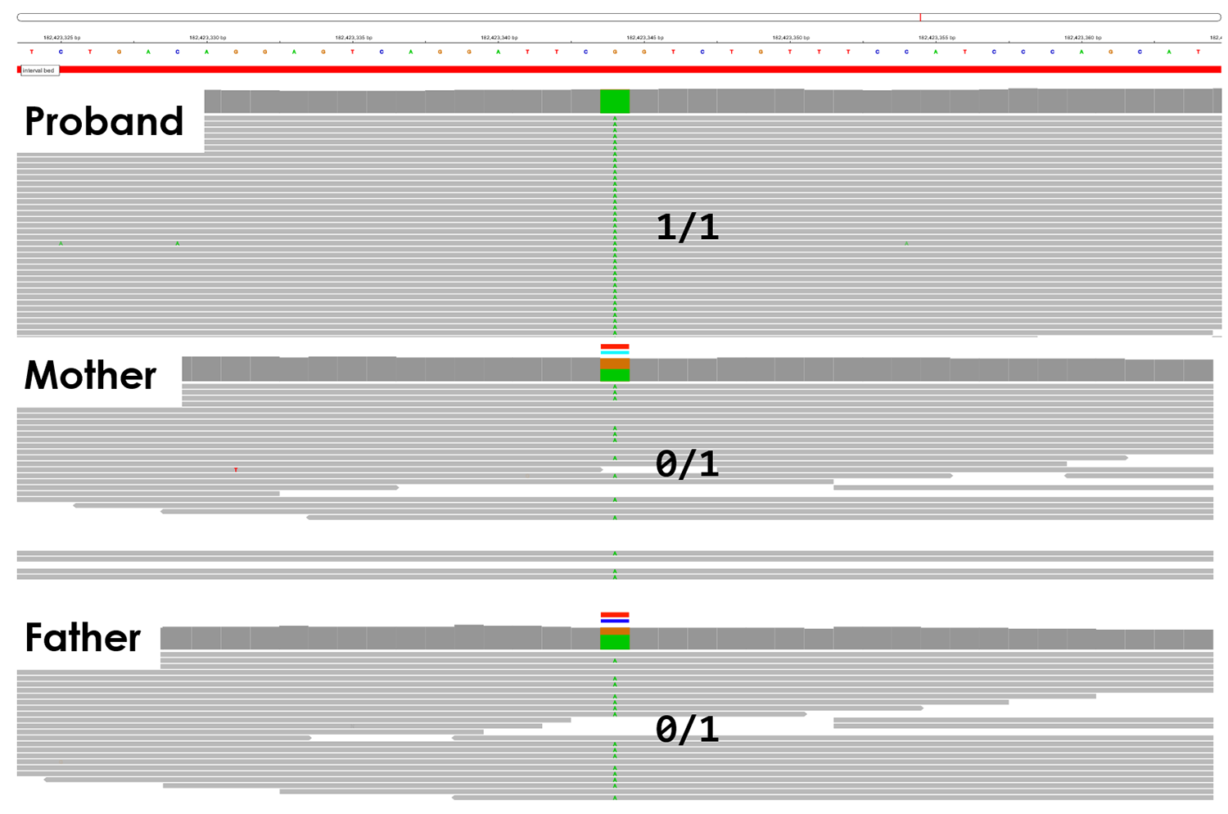
Google DeepTrio variant calling and Genoox Franklin Workbench interpretation identified a high-priority homozygous stop gain in the CERKL gene. In this case, both parents are heterozygous carriers. The variant is classified as pathogenic ClinVar and is associated with retinitis pigmentosa, a rare eye disease consistent with the patient phenotype. Pending confirmation via clinical sequencing, the variant is likely causal.
A Promising New Path to Democratize Trio Sequencing
This proof-of-concept study shows the AVITI system's promise for democratizing trio sequencing access via an affordable benchtop system and cloud-based analysis solutions. Much work remains to be done, and we are already pursuing more extensive studies across a broader range of disease contexts where de novo variants are more frequently responsible.
To hear more details about this study, watch Semyon Kruglyak, PhD, Vice President of Informatics at Element, present the team's results at AGBT below.