Avidity Base Chemistry
Breakthrough sequencing technology that resets expectations for cost, quality, and flexibility
Introduction to Element technology
Avidity base chemistry (ABC) brings a radically different solution to next-generation sequencing (NGS). Unique to Element, ABC reduces run costs and improves performance through innovations across all fronts of surface chemistry, biochemistry, engineering, and data analysis.
Low-bind surface chemistry
A low-binding surface chemistry captures template DNA on the surface of the flow cell for amplification and sequencing. Entirely new and extensible, the chemistry enables crisp, bright images that elevate the contrast‑to‑noise ratio (CNR) and improve base detection in all four channels.
Working in tandem with surface chemistry, an optical system featuring an extra-wide field of view images the flow cell with unprecedented range and remarkable edge-to-edge quality.
PCR-free amplification
Leveraging rolling circle amplification (RCA) for PCR-free amplification prevents template error propagation. In contrast to PCR-based amplification methods, such as bridge amplification, RCA replicates only the original DNA template, rolling each strand into a tightly bound polony consisting of many library copies in close proximity.
RCA not only provides high-fidelity amplification—it eliminates both optical duplicates and amplification-induced index hopping artifacts.
Rebuilt sequencing steps
ABC innovates each step of sequencing chemistry, most crucially separating base detection from strand extension to optimize enzyme conditions for each step to deliver unsurpassed accuracy by ensuring ultrapure base-calling signals while reducing costly reagent consumption.
Additionally, by forgoing modifier nucleotides and fluorescence cleaving, ABC generates scarless, natural DNA that confers superior performance over many cycles of sequencing.
Additional ABC advantages
ABC supports unique molecular identifiers (UMI), single- or dual-indexing, and single-read or paired-end runs to qualify a breadth of NGS capabilities on a single instrument:
- Short-insert sequencing
- Long-insert sequencing
- Synthetic long reads
- DNA sequencing
- RNA sequencing (RNA-Seq), including single cell
Learn more about ABC sequencing
Cloudbreak™ advancements
Cloudbreak is the latest advancement to ABC chemistry, scaling to suit any experiment and enhancing operational efficiency in multiple ways:
- Increase sequencing speed with 20% faster run times reduce 2 x 150 run times to < 40 hours and single cell experiments in less than a day.
- Load a linear library for automatic circularization onboard AVITI.
- Sequence and demultiplex Index 1 and Index 2 before Read 1 and Read 2 for real-time quality control and performance feedback.
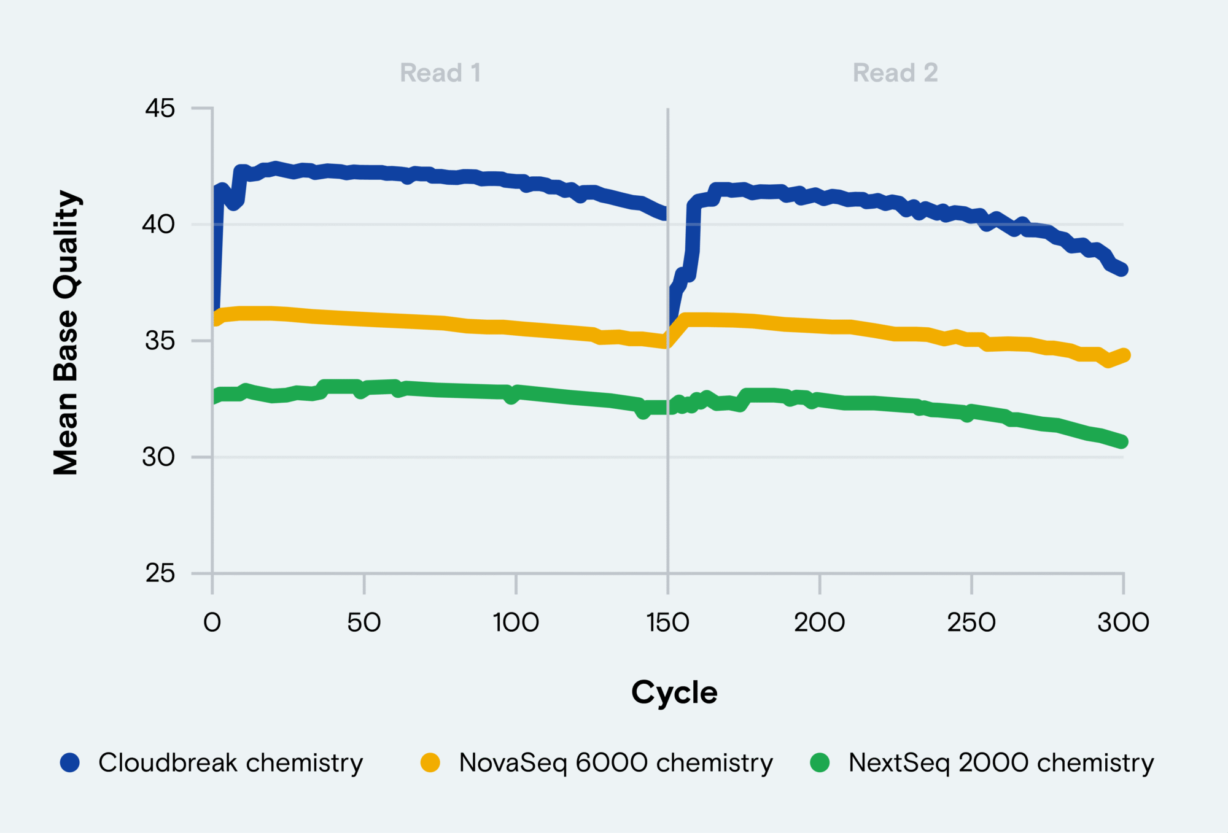
- Improve data quality through the end of each read to enhance accuracy.
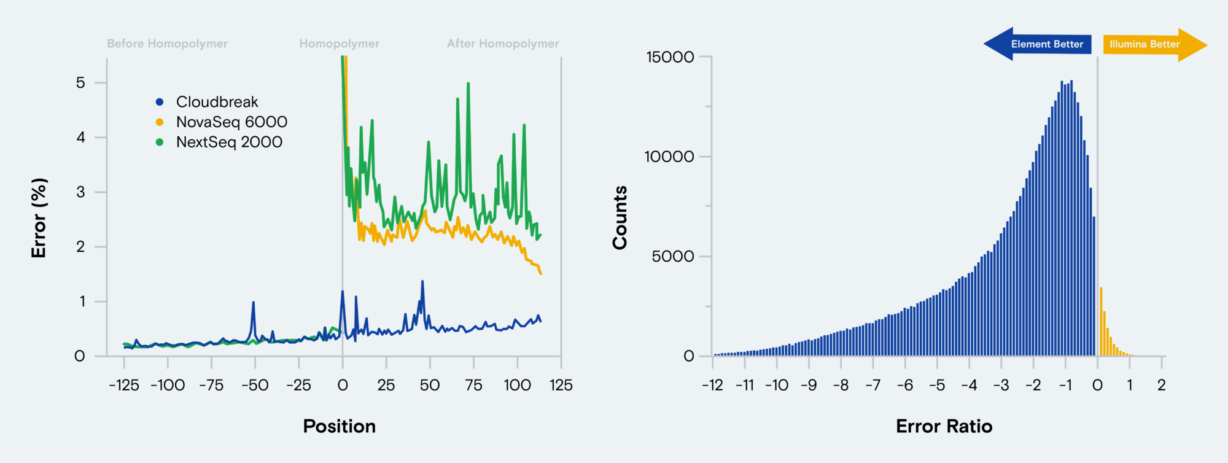
- Improve sequencing performance through difficult homopolymer regions.
Learn more about Cloudbreak kits
Complete sequencing ecosystem
The Element AVITI™ System grounds a prep-to-analysis sequencing portfolio built on ABC.
* Source: https://www.nature.com/articles/s41587-023-01750-7